Home Page – old draft
The goal of the lab is to reverse engineer neuronal activity to better understand and treat Neurodevelopmental disorders such as autism, epilepsy, and developmental delays. We combine computational (single and neuronal networks simulations) and experimental (patch-clamp Electrophysiology and multi-electrode arrays recordings) techniques.

Roy Ben-Shalom – Principal Investigator
twitter – @roybensh
Undergraduate Researchers:
Kyung Geun Kim – lorem ipsom
Alexander Ladd – lorem ipsom
Emily Nguyen– lorem ipsom
Jinan Liang — lorem ipsom
Michael Lam – – lorem ipso,
Using Deep Neural Networks to develop in silico neuronal models
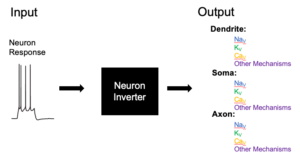
The goal of this project is to predict the biophysical properties of a neuron based on its electrophysiological response to stimuli. We built a deep learning convolutional network to predict the free parameters of a neuron model given it’s voltage response to a set of stimuli. Trainees in this project will have the opportunity to interact with various stages of the machine learning process, from data generation, analysis to neural network training. For this project we are looking for students with an interest in machine learning, optimization, statistics and/or neuroscience with good programming skills to help us improve the algorithm, generate training data, and increase the accuracy of our models Preprint
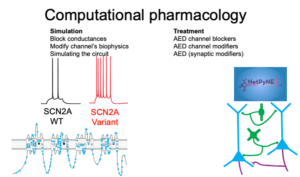
Simulate the effects of SCN2A variants on neuronal excitability and testing potential therapeutics
The SCN2A gene encodes the neuronal sodium channel Nav1.2, which is critical for the electrical activity of many types of neurons. Mutations in the gene alter the properties of the NaV1.2 channel, disrupting the function of single neurons and potentially leading to abnormal neuronal circuitry. Ultimately, these effects lead to epilepsy and/or ASD in affected patients. In this project, we will explore how alterations to the NaV1.2 channel affect the function of single neurons. Using computational models that simulate neuronal activity, we will study how each variant modulates single neuron activity and then simulate different drugs to see if they can reverse those effects.
Fitting neuronal models to Electrophysiological data.
Studying the biophysical properties of neurons, such as the different ion channel distributions and kinetics, can be done by fitting in silico neuronal models to in vitro neuronal recordings. In this process, a detailed neuron is modeled to a set of electrical circuits that describe its biophysical properties. The goal is to constrain the in-silico model’s parameters so it will behave like the in vitro neurons recorded during the experiment. This process is done using an optimization algorithm that relies on training the model across thousands of permutations.
We have developed a pipeline to fit the models to experimental data and we are now working on improving these models to develop highly detailed biophysical simulations for recorded neurons.

Electrophysiology and MEA recordings:
All the projects described above have experimental component to them which include empirical recording from neurons. We record from mouse acute slice, primary dissociated and iPSC derived neurons. Recordings are done using current/voltage -clamp MEA or combining current-clamp with MEA.
